The Landscape of WRKY-DNA Interactions
WRKY regulatory code database
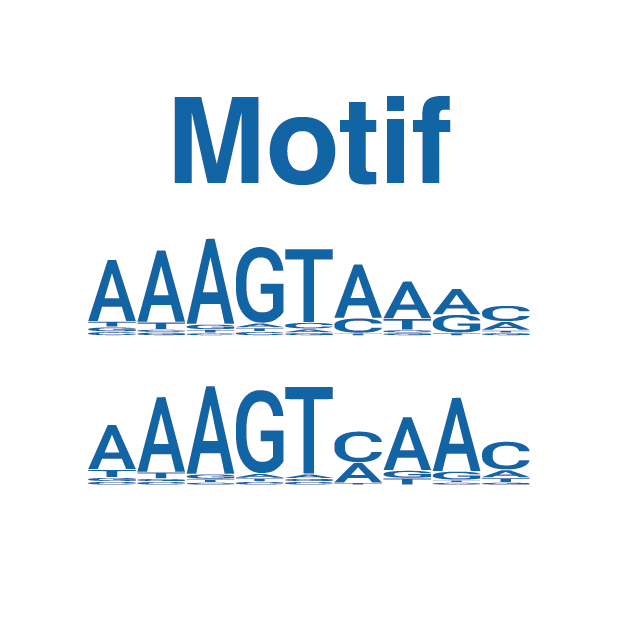
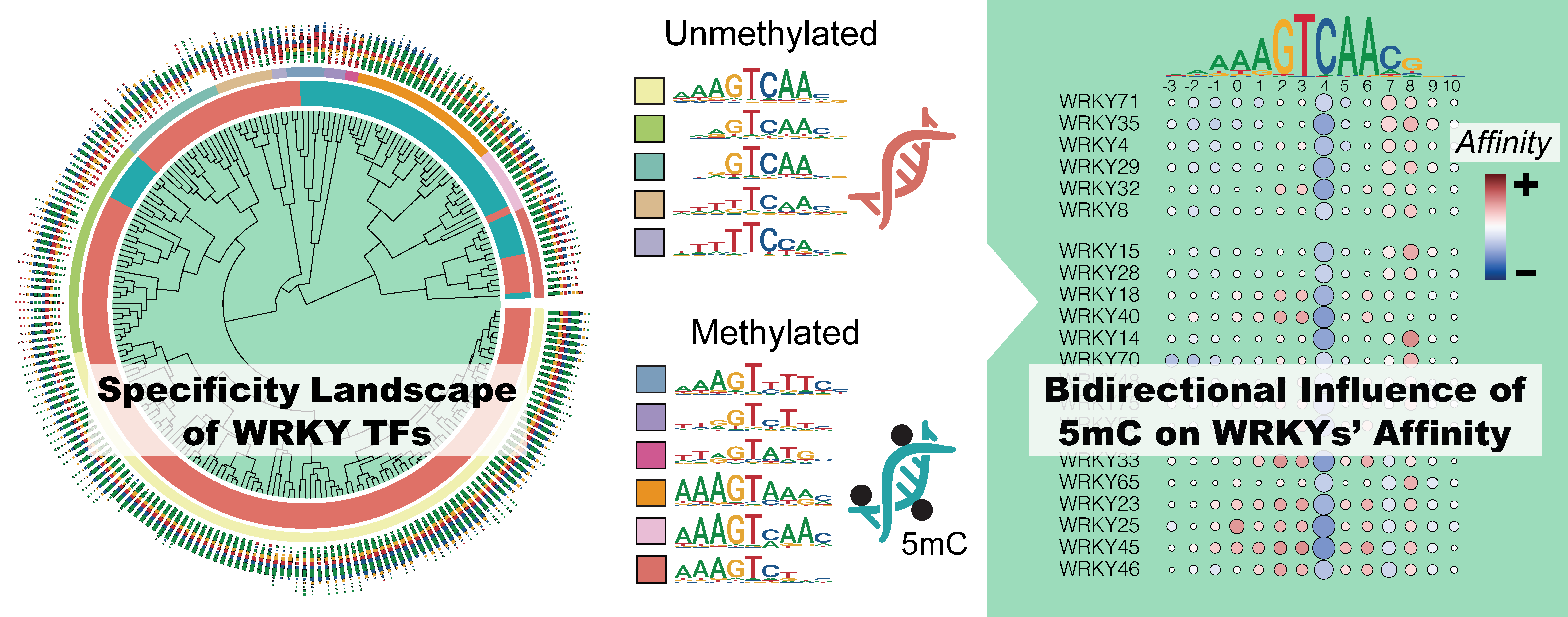
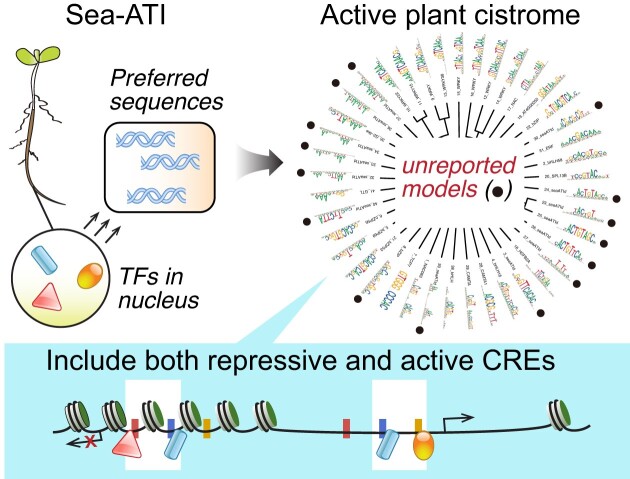
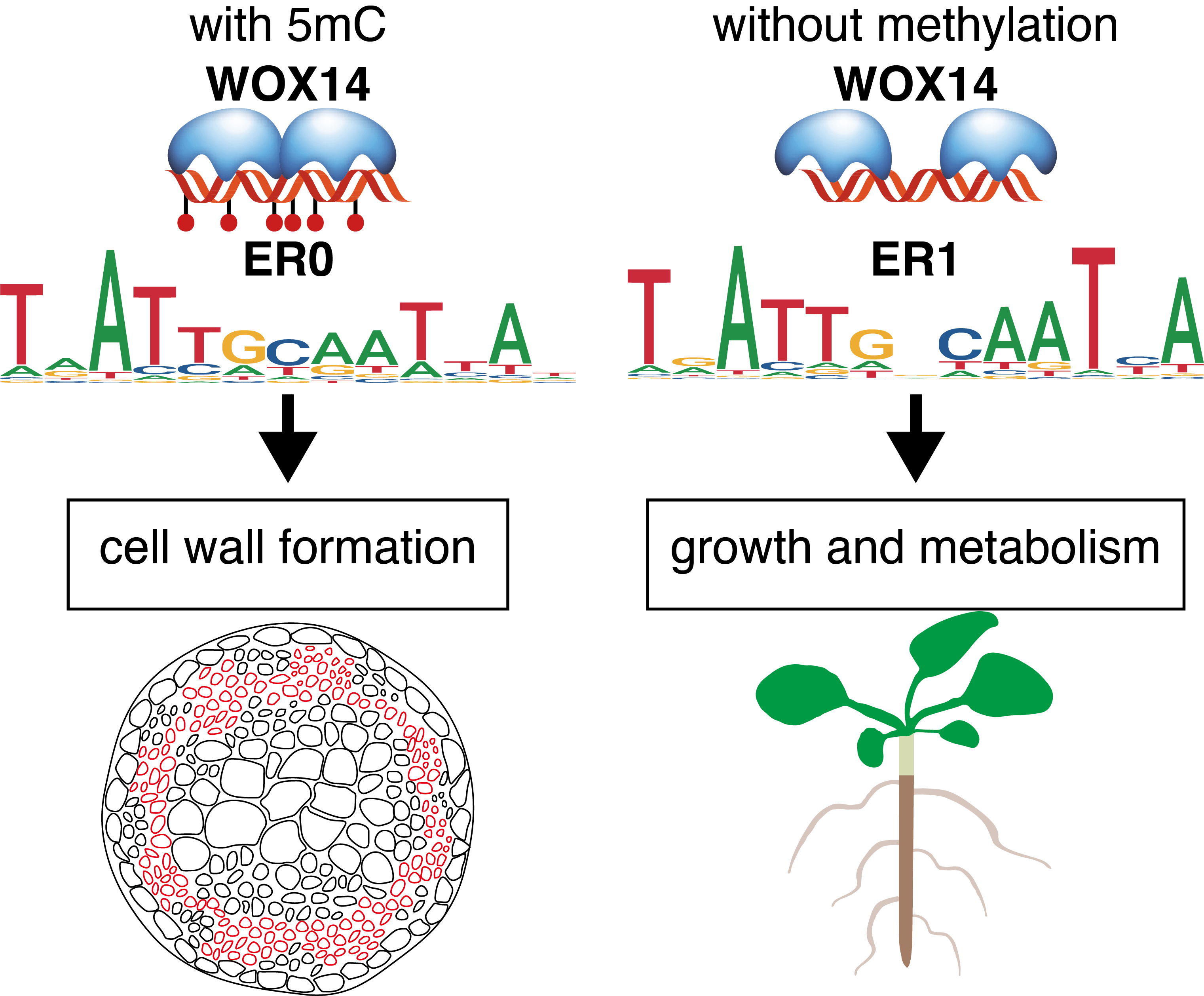
A total of 571 datasets, including SELEX, PBM, methyl-SELEX, DAP, ampDAP, methyl-ampDAP, and ChIP-seq data, were curated and analyzed to depict the TF-DNA interaction landscape of AtWRKYs both in the presence and absence of DNA methylation.
202
DAP
DAP
120
ampDAP
ampDAP
65
methyl-ampDAP
methyl-ampDAP
129
SELEX
SELEX
39
methyl-SELEX
methyl-SELEX
16
ChIP
ChIP